(Updated 22nd march 2021: added datasource.ai)
Here is, to my knowledge, the most complete list of data science competition platforms with sponsored (paid) competitions.
If you are not familiar with them, they are, to me, the best way to learn data science. Most of them have a dedicated community and many tutorials, starter kits for competitions. They are also a way to use your skills on topics your day job may not propose you.
If I forgot any platform, or if a link is dead, please let me know in the comments or via email, I plan to keep this article as updated as possible!
ML contests: an aggregator
Clear. A list of competitions, by topics (NLP / supervised learning / vision …)
I am not sure all the platforms are showed here (I did not find references to numer.ai or Bitgrit, per example)
Prediction
The principle here is simple, you have a train set and a test set to download (though the new trend is encouraging to push your code directly in a dedicated environment hosted by the platform).
The train set contains various columns, or images, or executables (or anything else probably) and the purpose is to predict another variable (which can be a label for classification problems, a value for regression, a set of labels for multiclass classification problem or other things, I am just focusing on the most common tasks)
Then, you upload your predictions (or your code, depending on the competition) and you get a value: the accuracy of your model on the test set. The ranking is immediate, making these platforms delightfully and dangerously addictive!
Kaggle
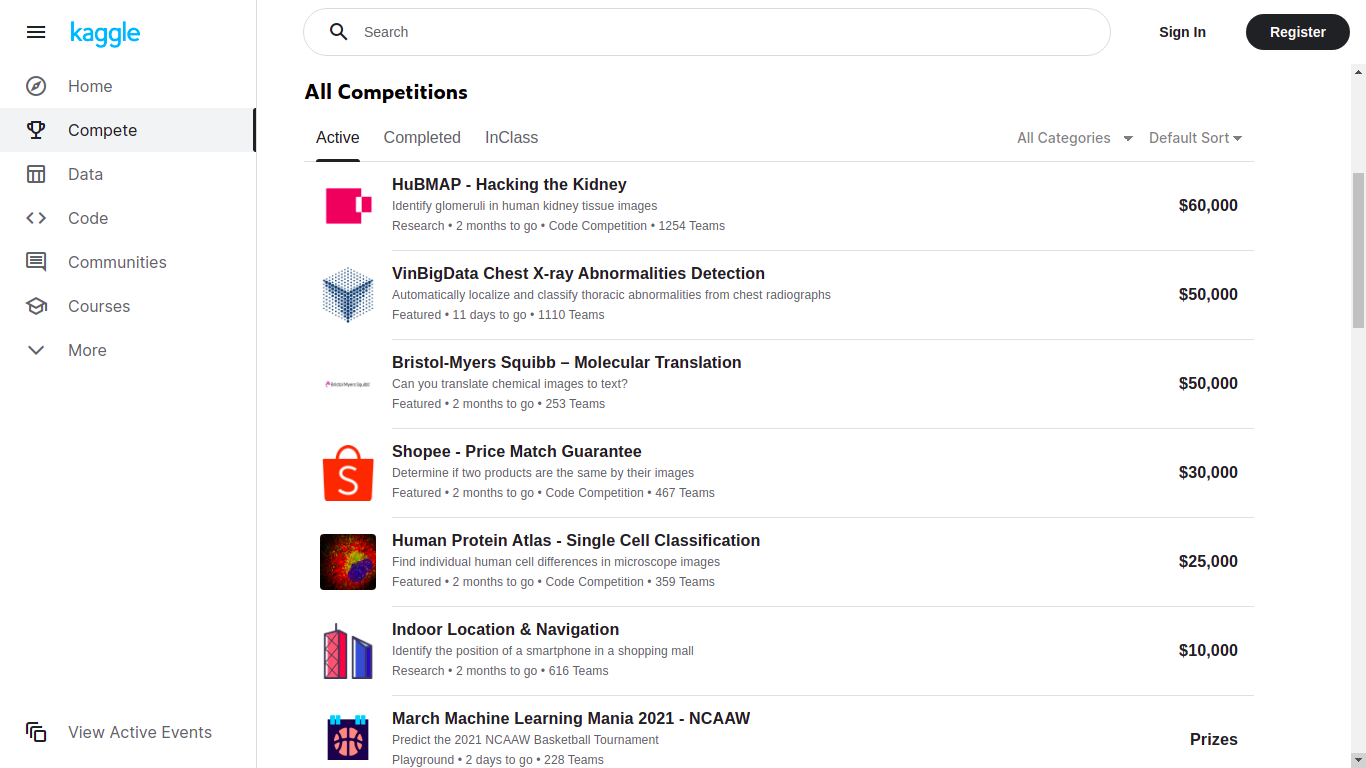
Kaggle is probably the largest platform hosting competitions, with the highest prizes and the largest community and resources. Beware, the higher the price, the harder the competition!
They also have the most complete set of learning resources and usable datasets.
AIcrowd (or CrowdAI)
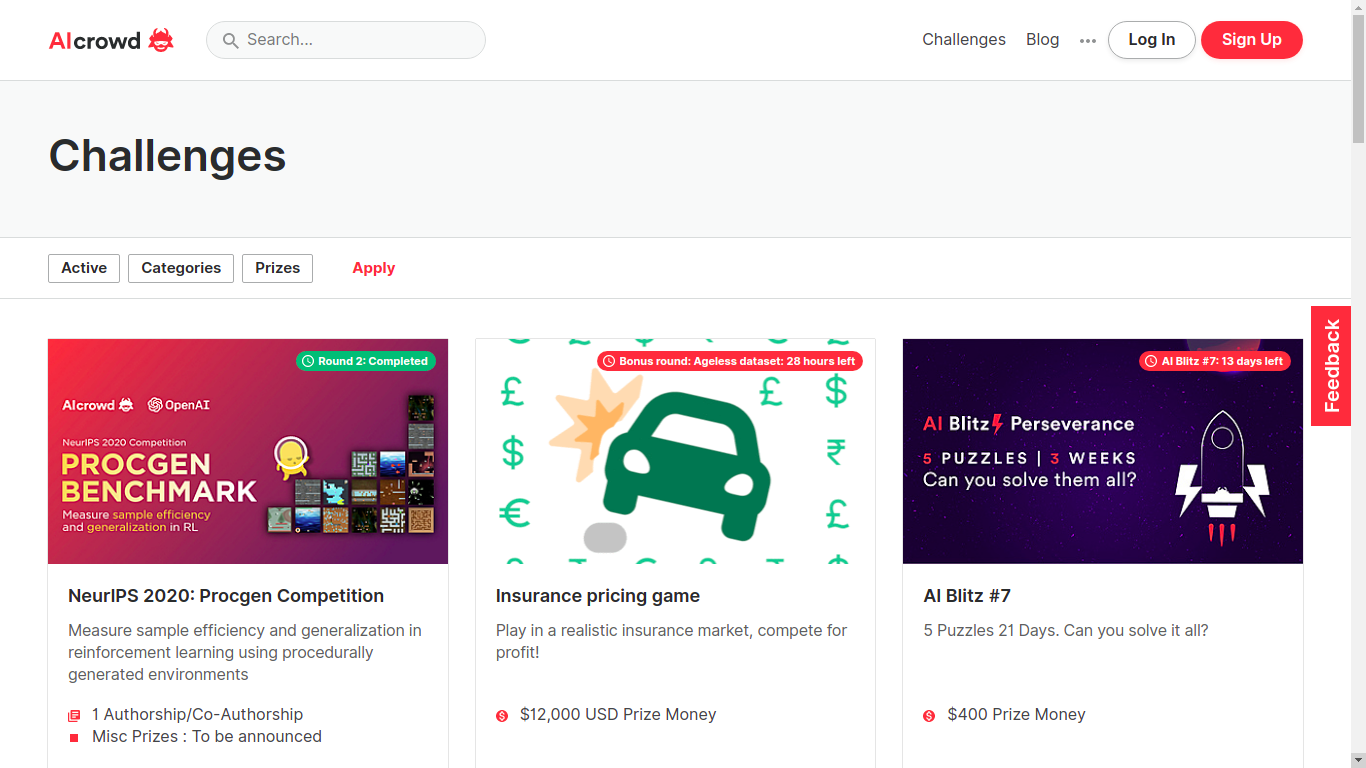
Great platform, super active, many competitions and great topics! They are growing fast so expect even more competitions to happen here.
Besides, from the competitions I have seen here, they focus on less “classical” topics than the ones you would see on Kaggle. Some may like it, others may not, I personnally do.
AIcrowd enables data science experts and enthusiasts to collaboratively solve real-world problems, through challenges.
Bitgrit
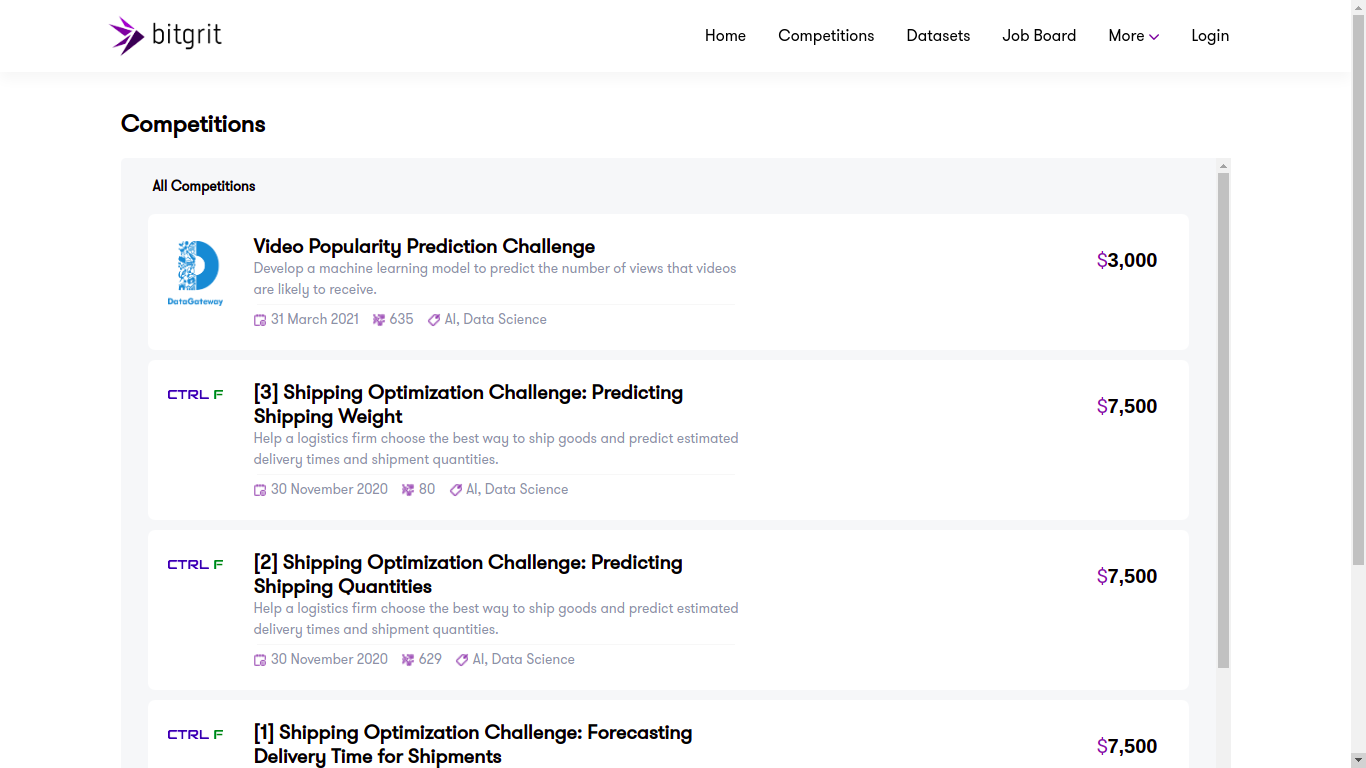
Launched in 2019, already showing 8 competitions with various topics, this platform looks promising! As said above, it does not seem referenced on mlcontests.
bitgrit is an AI competition and recruiting platform for data scientists, home to a community of over 25,000 engineers worldwide. We are developing bitgrit to be a comprehensive online ecosystem, centered around a blockchain-powered AI Marketplace.
Drivendata
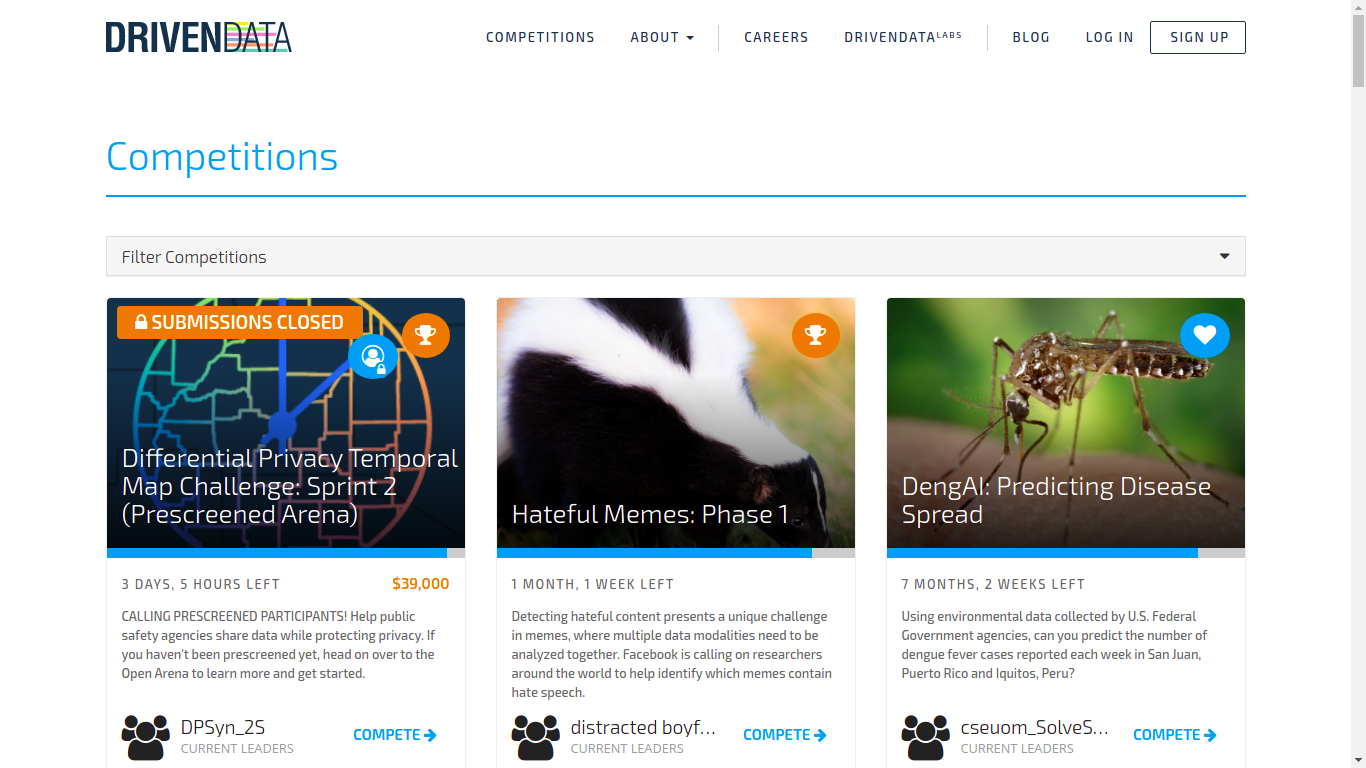
I never took part in their competitions, so I can’t say mcuh about it for now! But they have sponsored competitions.
DrivenData works on projects at the intersection of data science and social impact, in areas like international development, health, education, research and conservation, and public services. We want to give more organizations access to the capabilities of data science, and engage more data scientists with social challenges where their skills can make a difference.
Crowdanalytix
I never took part in their competitions, so I can’t say mcuh about it for now! They seemed less active recently, but they had sponsored competitions.
25,129 + Data Scientists
102,083 + Models Built
50 + Countries
Numer.ai
Focusing on predicting the stock market, with high quality data (which is usually a tedious task when you try to have quality data in finance). They claim to be the hardest platform in finance, and having worked there, I can confirm that finding the slightest valuable prediction is super hard!
Nice if you like finance, but be prepared to work with similar datasets!
Start with hedge fund quality data. It is clean and regularized, designed to be usable right away.
Zindi
Data science platform with competitions which are related to Africa. The NLP part seems particularly exciting, as they are focus on languages which are not studied as often as English or Spanish! Looking forward to participate in one of their challenges!
We connect organisations with our thriving African data science community to solve the world’s most pressing challenges using machine learning and AI.
Analytics Vidhya
India based.
Data science hackathons on DataHack enable you to compete with leading data scientists and machine learning experts in the world. This is your chance to work on real life data science problems, improve your skill set, learn from expert data science and machine learning professionals, and hack your way to the top of the hackathon leaderboard! You also stand a chance to win prizes and get a job at your dream data science company.
Challengedata

Not sure the competitions are sponsored here. General topics, most of them seem to come from French companies and French institutions.
We organize challenges of data sciences from data provided by public services, companies and laboratories: general documentation and FAQ. The prize ceremony is in February at the College de France.
Coda Lab
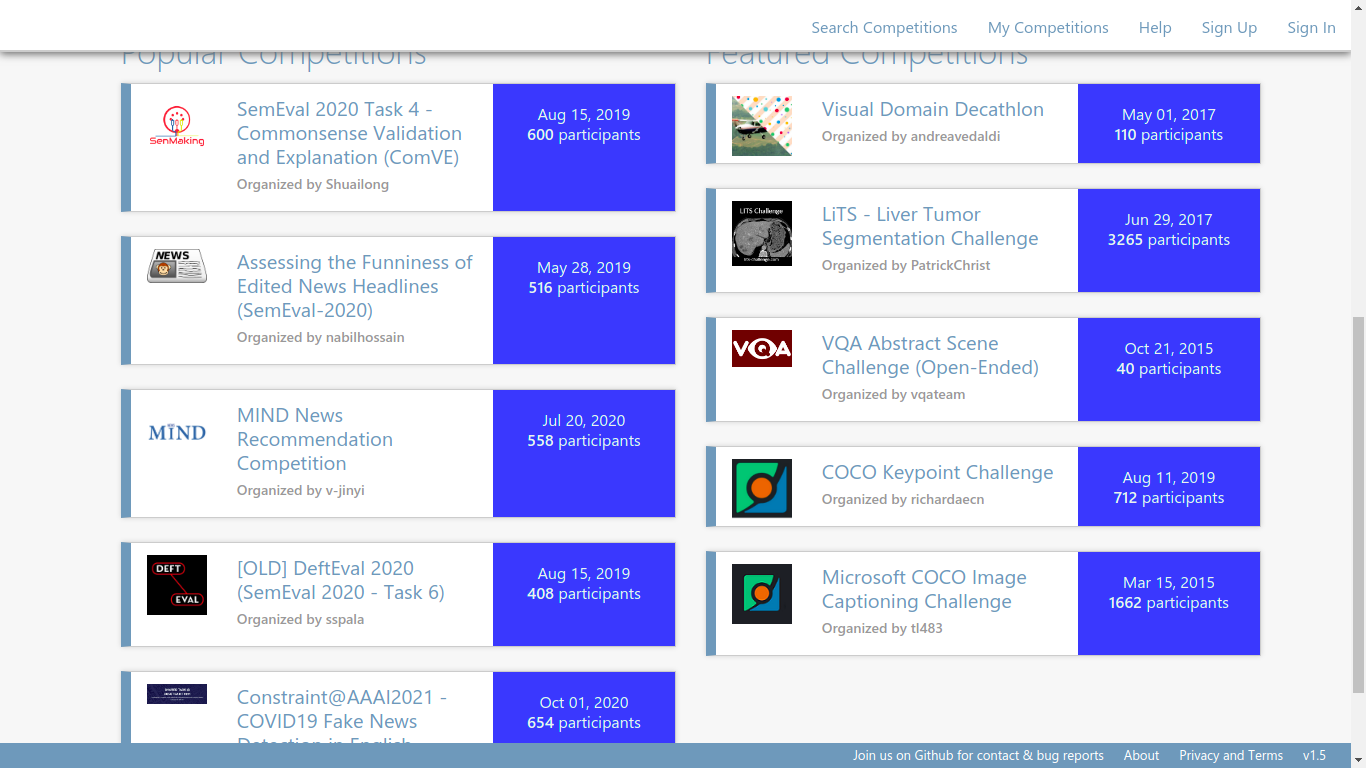
French based.
CodaLab is an open-source platform that provides an ecosystem for conducting computational research in a more efficient, reproducible, and collaborative manner.
Topcoder

Not focusing only on data science:
Access our community of world class developers, great designers, data science geniuses and QA experts for the best results
InnoCentive
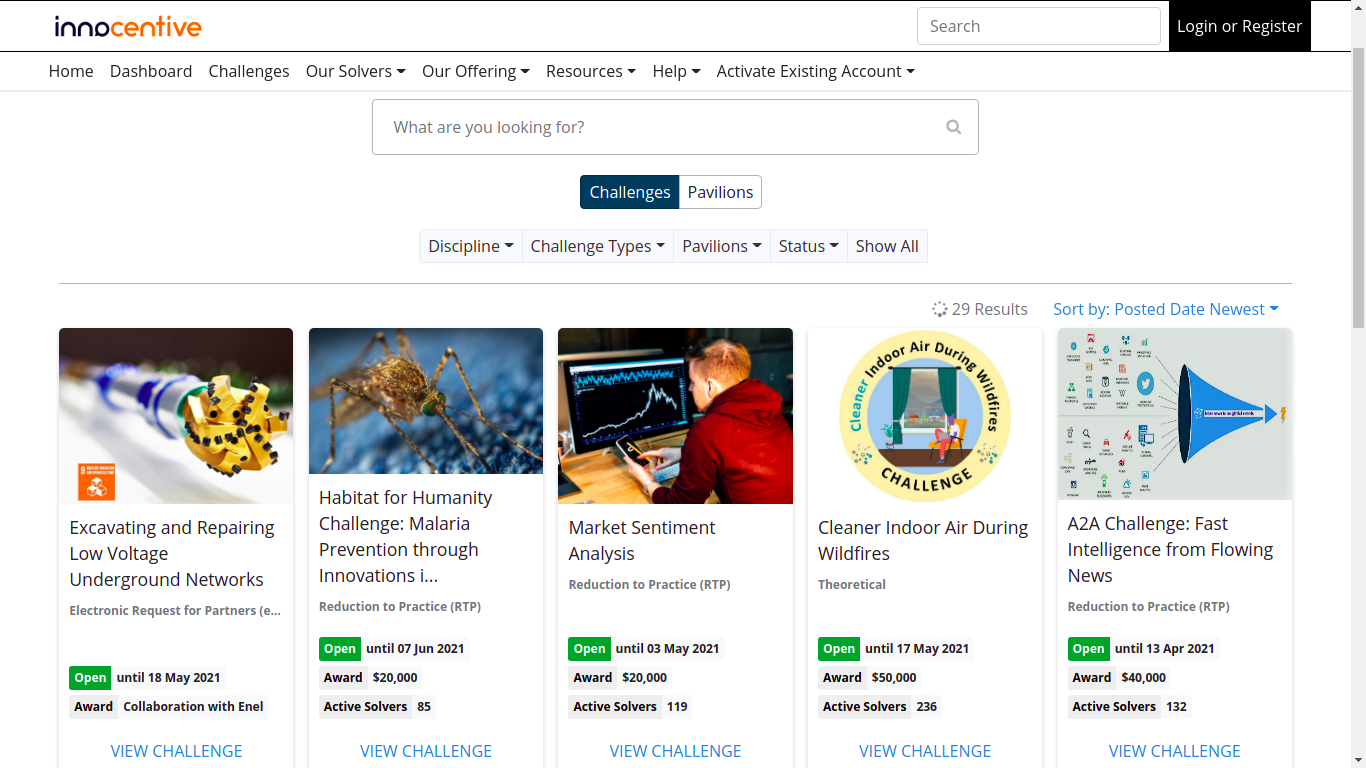
InnoCentive is the global pioneer in crowdsourced innovation. We help innovative organizations solve their important technology, science, business, A/I and data challenges by connecting them with a global network of expert problem solvers.
Datasoure.ai
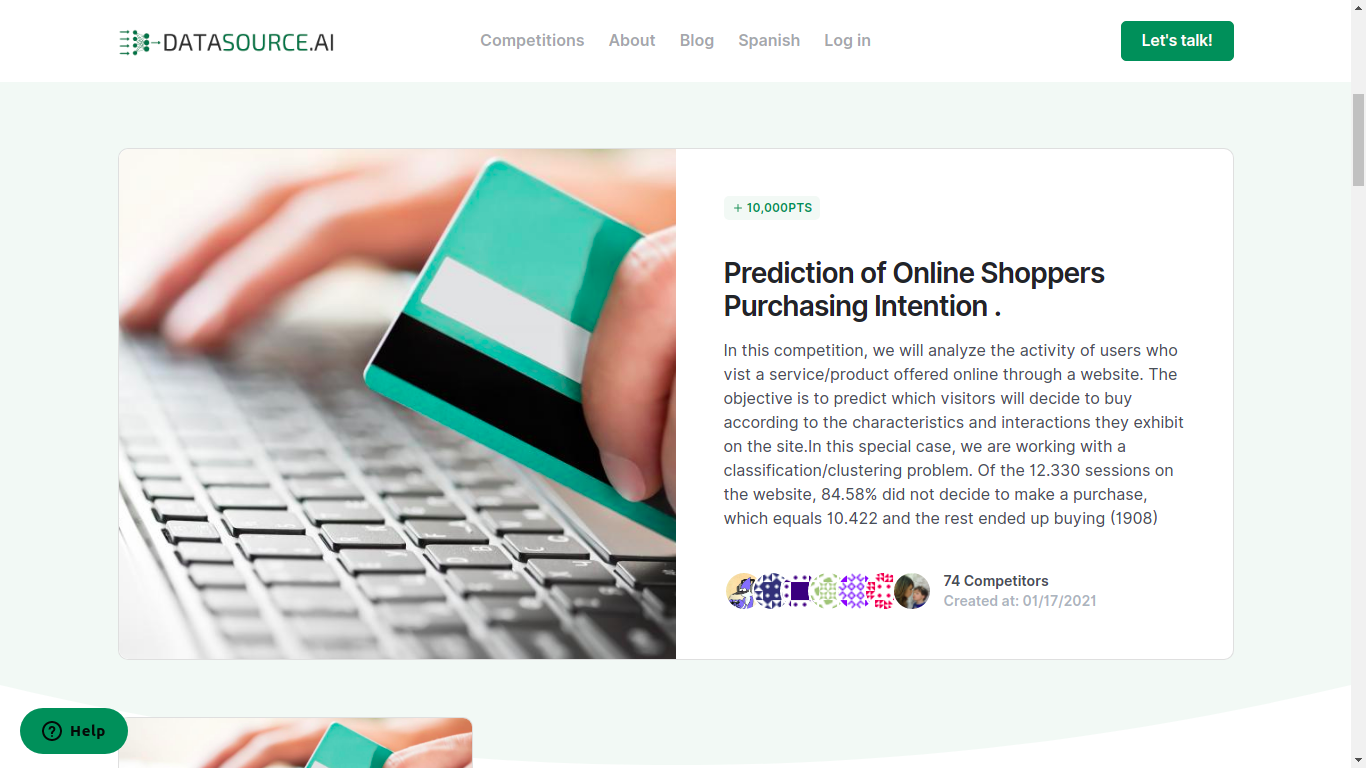
Young company, as the quote below shows (22nd march 2021). They seem to be focused on challenges for startups, but this may evolve!
At a glance
2 Team Members
1,692 Data Scientists
12 Companies
5.2% Weekly Growth
Signate
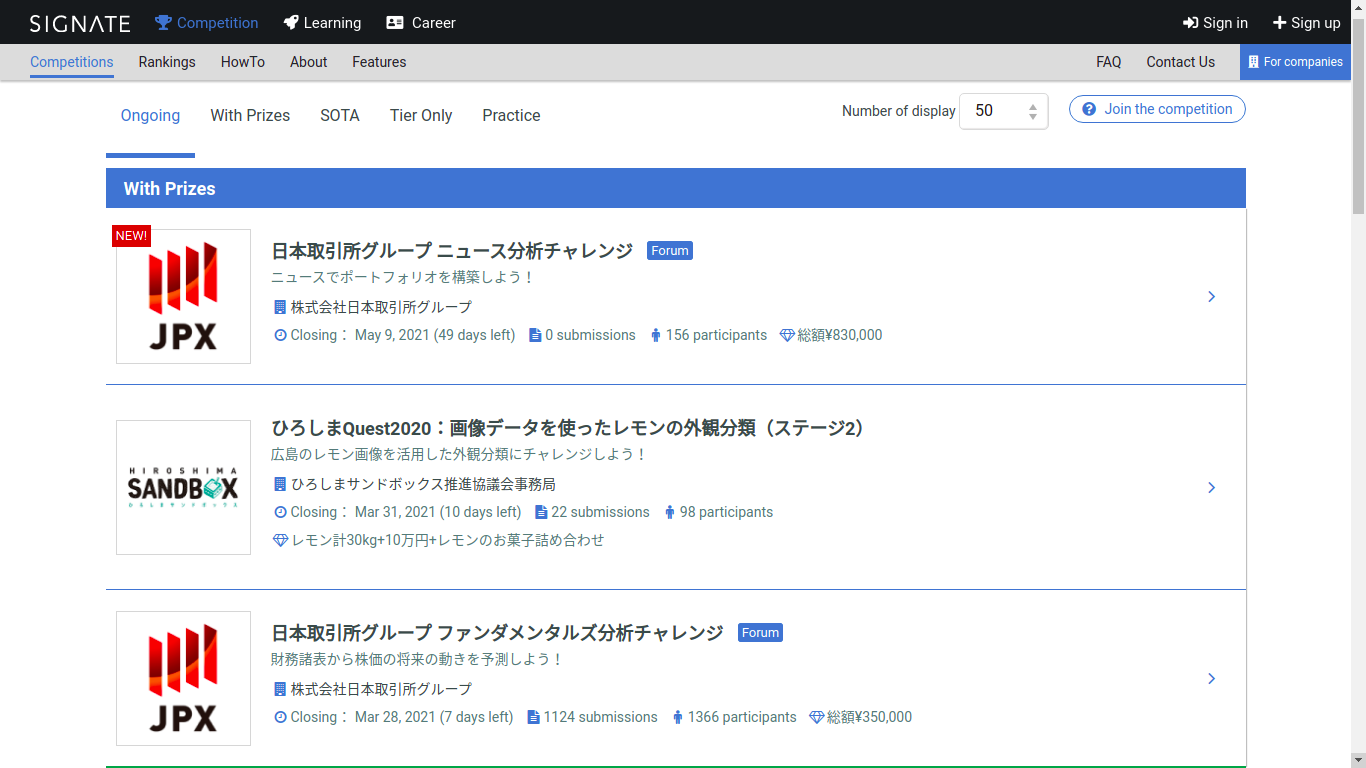
A Japanese competition platform. Most of the competitions are described in Japanese, but not all of them!
SIGNATE collaborates with companies, government agencies and research institutes in various industries to work on various projects to resolve social issues. We invite you to join SIGNATE’s project, which aims to make the world a better place through the power of open innovation.
datasciencechallenge.org (probably down)
https://www.datasciencechallenge.org/
Unfortunately, I cannot reach the website any more…
Sponsored by the Defence Science and Technology Laboratory and other UK government departments.
datascience.net (probably down for ever)
Used to be a French speaking data science competition for a while. However, the site has been down for a while now… Worth giving a look from time to time!
Dataviz
Here, the idea is to provide the best vizualisation of datasets. The metric may therefore not be as absolute as the one for prediction problems and the skillset is really different!
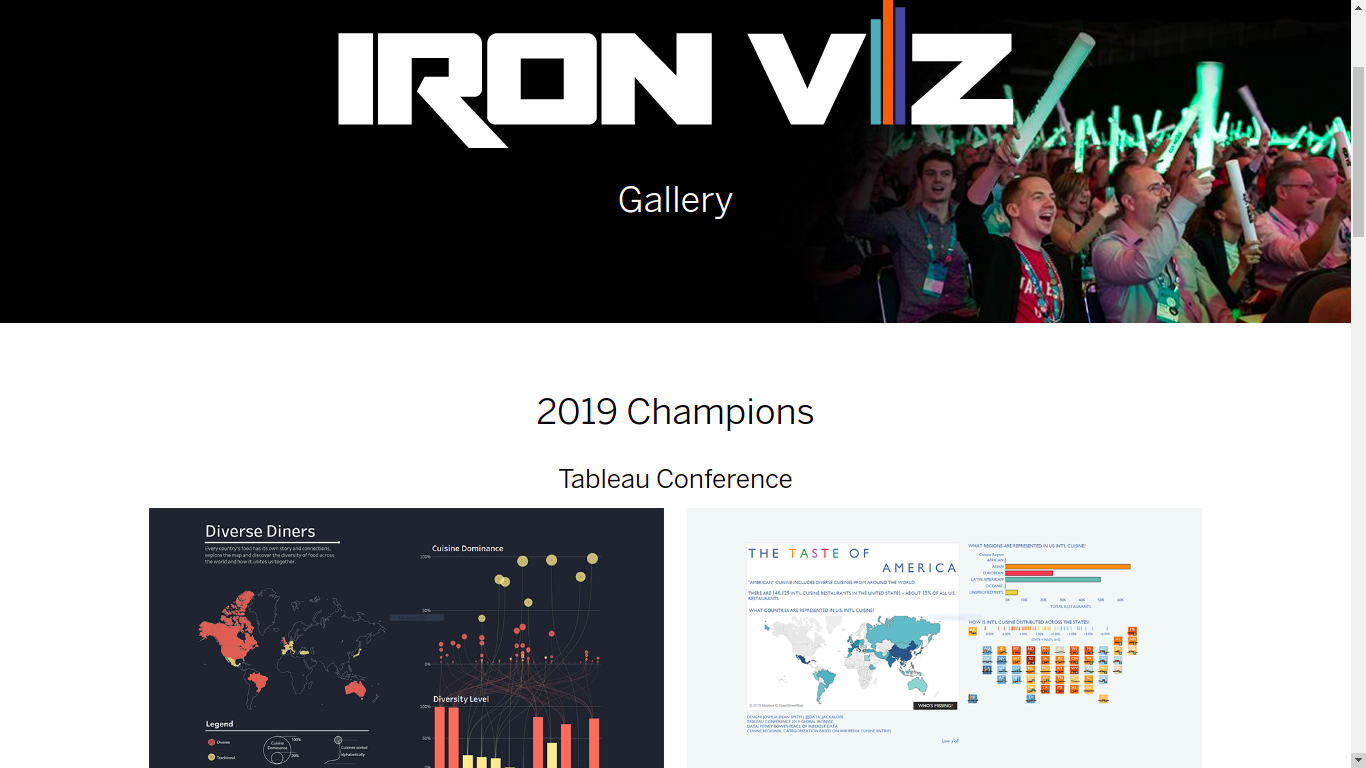
Iron viz
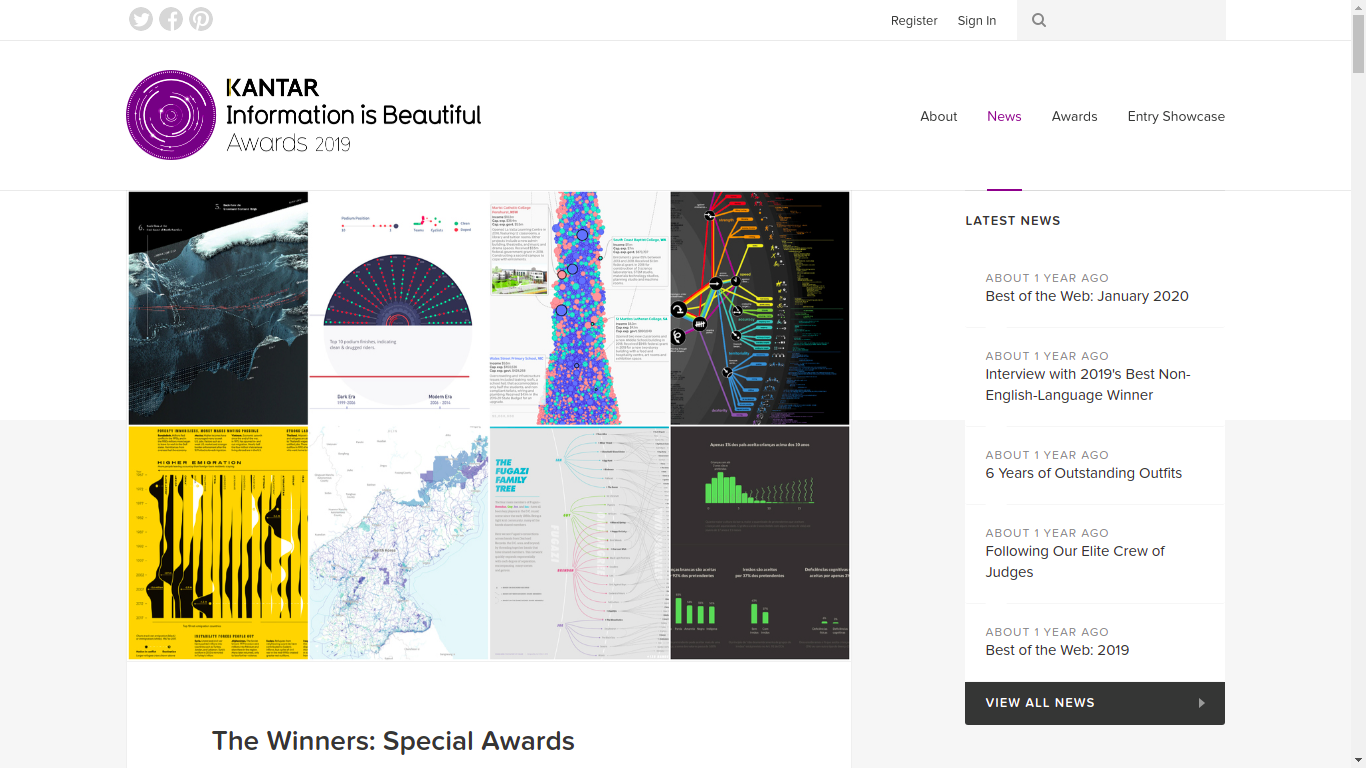
informationisbeautifulawards
They are all the platforms I am aware of, if I missed any or if you have any relevant resources, please let me know!
I Hope you liked this article! If you plan to take part in any of these competitions, best of luck to you, and have fun competing and learning!




_Contour.png)
_Contour.png)